Optogenetics and calcium imaging have advanced the field of neuroscience by providing precise methods for manipulating and imaging neural activity in vivo. In order to map large-scale cortical circuits in a head-fixed animal, it is essential to have the ability to perform large-scale targeted optogenetics and imaging in vivo. Below you can find application examples.
Targeted Optogenetic Stimulation of Motor and Non-Motor Cortices
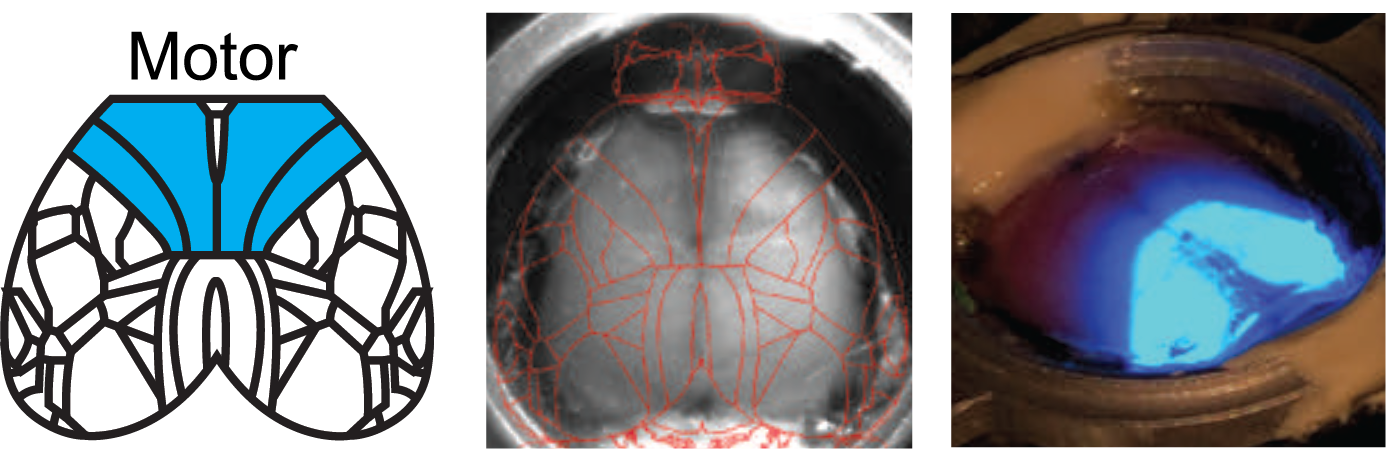
Kauvar et al. used the Mightex Polygon to optogenetically inhibit the activity of multiple cortical regions—at the same time—in awake mice. They designed a calibration system to align the illumination to specific regions of cortex, and could synchronize illumination to different parts of a task the mouse was performing. This calibration system is demonstrated in these images. The Mightex system enabled them to target nearly the entirety of dorsal cortex. This causal manipulation was paired with another technique that we developed, COSMOS (see https://sites.google.com/view/getcosmos), which enables simultaneous recording of neural activity across the extent of mouse dorsal cortex. Using COSMOS, they found that there is activity throughout cortex related to the mouse’s task performance; using the Polygon, we could then show that inhibition of multiple cortical regions could disrupt task performance.
Cortex-Wide Optogenetics
This video shows our OASIS Macro and laser Polygon DMD illuminator being used to target ChR2 optogenetic stimulation (200um spots) across the cortex of an anesthetized mouse through a cranial window. This experiment was performed to elicit paw motor movements (Courtesy of University of Ottawa).
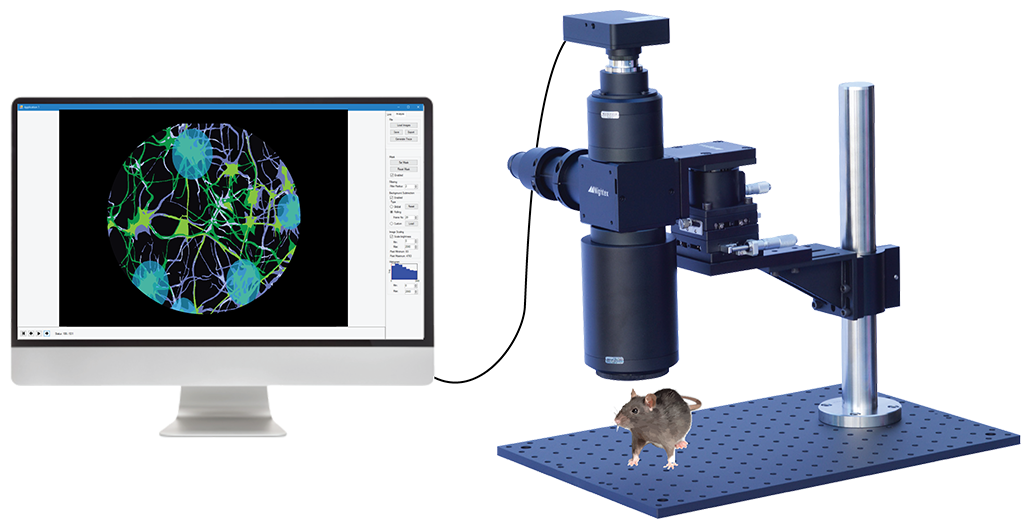
Mesoscopic Imaging and Optogenetics
with Mightex’s OASIS Macro
OASIS Macro Inquiry
"*" indicates required fields