Published on 2023/07/11 Research powered by Mightex’s Polygon1000 
 Honda, M., Oki, S., Kimura, R., Harada, A., Maehara, K., Tanaka, K. & Ohkawa, Y., High-depth spatial transcriptome analysis by photo-isolation chemistry. Nature Communications,12(1), 4416 (2021).
Honda, M., Oki, S., Kimura, R., Harada, A., Maehara, K., Tanaka, K. & Ohkawa, Y., High-depth spatial transcriptome analysis by photo-isolation chemistry. Nature Communications,12(1), 4416 (2021).

Discover the cutting-edge technique by Honda et al. (2021) that uses the Mightex Polygon for targeted gene expression analysis in tissues with multiple cell types. Their method, detailed in Nature Communications, precisely profiles regions of interest by photo-uncaging primers with the Polygon, allowing for high-resolution transcriptomic insights of specific cells or structures. This advancement opens new doors in the realm of deep expression profiling, showcasing the versatility of the Mightex Polygon in complex tissue analysis.
Introduction
Most tissues aren’t composed of a single cell type – for example, renal glomuruli are composed of many cell types: mesengial cells, endothelial cells, podocytes, and more. If a researcher wanted to better understand the current gene expression of endothelial cells in renal glomuruli, how might they do it? There is no easy way to separate the transcription profile of mesengial cells, endothelial cells, and podocytes. For the purpose of deep expression profiling of tissue with multiple cell types, Honda et al. (2021) have developed a novel technique that allows researchers to profile specific regions of interest (ROI) in a given tissue.
Methods & Findings
In this technique, the tissue of interest is infused with photo-caged primers and reverse transcriptase. Next, a digital micromirror device ( Polygon 400, Mightex ) is used to illuminate precise patterns on the tissue with UV light, uncaging only those primers that lay in the ROI (Fig 1A). Following this, the tissue is lysed and cDNA is collected and sequenced using CEL-seq2 (Hashimshony et al., 2016). Using this technique, only those cells which lay in the ROI have their transcriptome’s profiled. This can be used to characterize specific cell types, or to characterize distinct structures (Fig 1B).
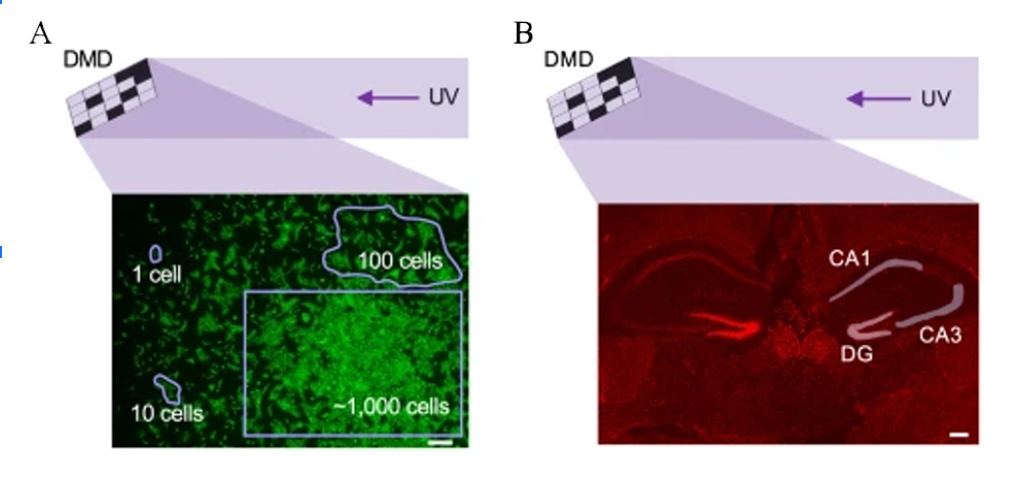
Figure 1: Visualization of a digital micromirror device being used to illuminate different types of ROI. The Polygon, the device used by Honda et al., can precisely illuminate structures with sub-micron resolution. A) Examples of a range of ROI shapes that can be created using the Polygon (Mightex). B) ROI illumination patterns being used to investigate the hippocampal gene expression.
Findings and Conclusion
In their paper, Honda et al. (2021) discuss two distinct example use cases for this technique. In the first, the researchers show that you can target a specific cell type, and have each cell of that type be an ROI. To do this, the cell-type of interest is first immunistained using antibodies that target cell-type specific markers. The tissue can then be viewed under a fluorescent microscope and the fluorescently labelled cells can each become an ROI. The second application is for stimulating larger structures. In this case, bitmap image masks were created in the shape of the CA1, CA3, and dentate gyrus (DG). These bitmaps were uploaded to the DMD (Polygon, Mightex) to create the three distinct ROI.
One factor in effectively isolating spatially defined regions is ensuring that the contrast between the illuminated ROI and the unilluminated regions is high enough that the primers in the unilluminated regions don’t become uncaged. Importantly, Honda et al. (2021) found that, after optimizing their protocol, there weren’t any detectable background signals from non-irradiated cells.
While many distinct expression profiling techniques have been developed in recent years, deep expression profiling continues to be difficult, especially when profiling specific structures or small numbers of cells. This new tool presented by Honda et al. allows researchers to use photo-isolation chemistry to profile specific cell types and structures with a high degree of precision.
Joe Banning, Applications Specialist at Mightex
To read the full publication, please click here.



