
Light-Seq: light-directed in situ barcoding of biomolecules
in fixed cells and tissues for spatially indexed sequencing
Published on 2022/11/06 Research powered by Mightex’s Polygon1000 



Check out this recent publication using the Polygon1000! The paper outlines research by Jocelyn Kishi, Ninning Liu, Emma R. West, and colleagues from Harvard University using the Polygon for UV photocross-linking for a novel method of DNA barcoding. We are excited to see more to come using the LightSeq technique!
Introduction
Cellular biologists are often interested in visualizing the properties of cells within a particular sample, and sorting them based on particular features such as physiological properties or spatial location. In order to reach this goal, scientists often employ opto- or microfluidic approaches before, during, or after imaging the sample. Alternatively, photo-conversion or photo-activation methods provide fluorescent tags on particular cells of interest that can subsequently be visualized. These existing cell sorting techniques have some disadvantages as they usually require aspiration of live cells for imaging purposes, and for single cell sorting post-imaging. However, this often results in the destruction of the tissue sample and valuable information, such as spatial location, overlap of cellular processes, and other nuanced morphological features can be lost.
In order to overcome this limitation, sequencing techniques were developed for DNA barcoding, to spatially index two-dimensional surfaces. These barcodes can then be read out in situ via subsequent iterative imaging processes or ex situ by post-retrieval sequencing. These techniques require several pieces of expensive and highly specialized equipment and experimental protocols are severely limited by technical capabilities of this equipment, often resulting in sub-optimal results and long experimental timelines. As such, Kishi and colleagues sought to develop a novel method of DNA bar coding for cell sorting that allows tissue samples to remain intact while employing low cost and versatile equipment to produce high throughput results.
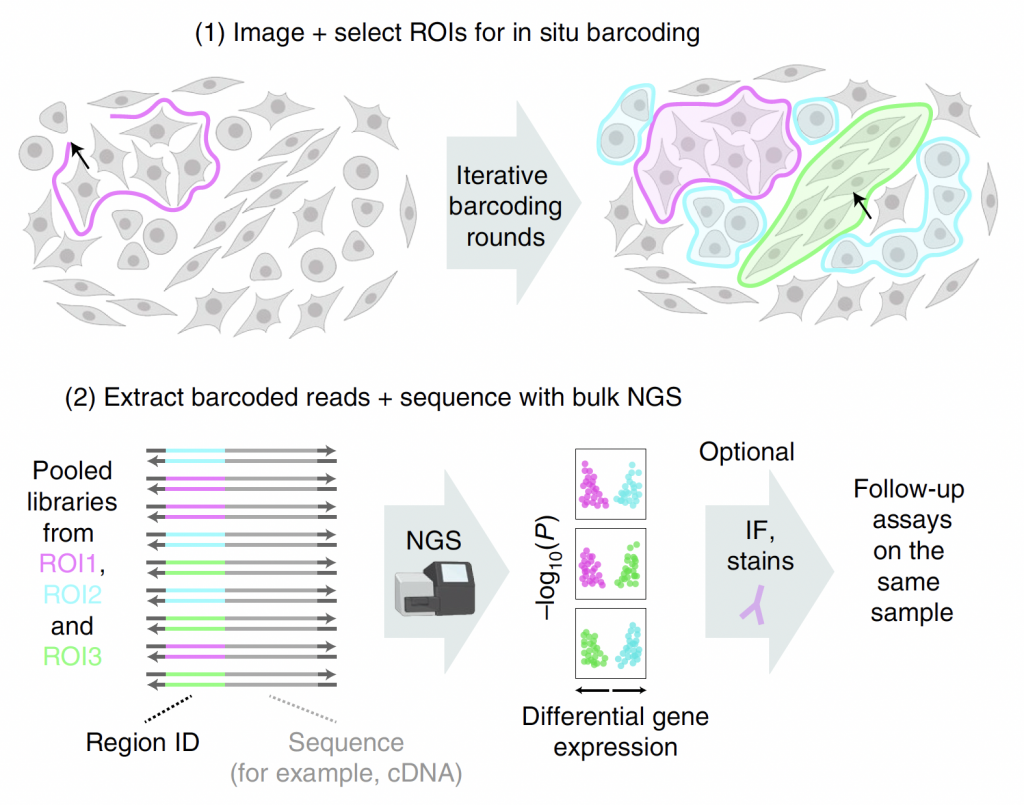 Figure 1: Adapted from Source. Selective barcoding of custom selected cells or tissue regions. Panel 1. definition of ROIs, based on phenotypic factors post-imaging. This can be completed manually or automatically.Panel 2. Barcode generation and expression via photocrosslinking with patterned UV illumination targeted selectively at ROIs.
Figure 1: Adapted from Source. Selective barcoding of custom selected cells or tissue regions. Panel 1. definition of ROIs, based on phenotypic factors post-imaging. This can be completed manually or automatically.Panel 2. Barcode generation and expression via photocrosslinking with patterned UV illumination targeted selectively at ROIs.
Methodology
In this paper, Kishi and colleagues outline the light controlled DNA barcode attachment strategy using imaged examples. They use barcode strands containing the ultrafast photocrosslink 3-cyanovinylcarbazole nucleoside (CNVK)31 that is able to form an interstrand crosslink with application of UV illumination.
By first determining the cells of interest and defining regions of interest (ROIs) for each area for photocrosslinking, Kishi and colleagues were able to target their crosslinking to specific cellular populations. That is, the authors were able to illuminate a group of ROIs with UV light in the presence of complementary docking sequences to label with a particular fluorescent tag. Multiple ROIs can be labelled with the same tag at once and multiple populations can be tagged overtime with temporally distinct patterned illumination.
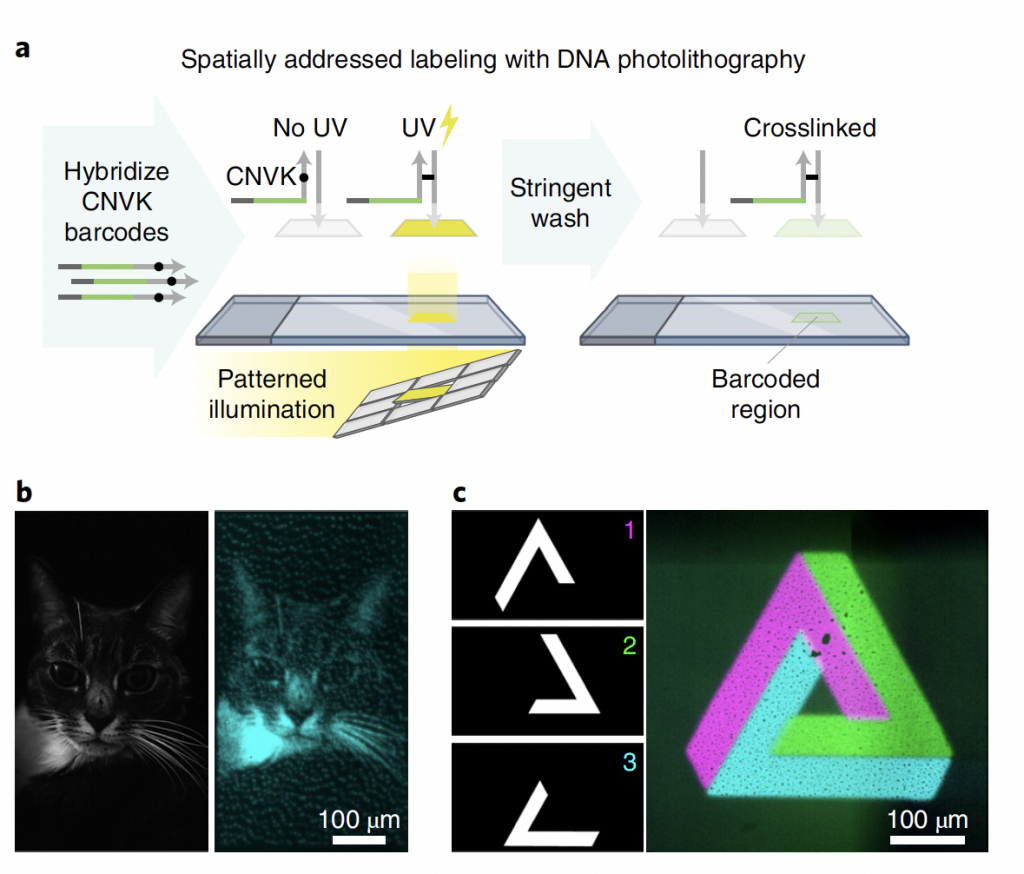 Figure 2: Adapted from Source. Application of Patterned Illumination for photocrosslinking with UV illumination Panel A. Experimental steps for photocross-linking Panel 2. UV photocross-linked examples of fluorescent barcode strands (left- single coloured cat; right 3 colour orthogonal shapes)
Figure 2: Adapted from Source. Application of Patterned Illumination for photocrosslinking with UV illumination Panel A. Experimental steps for photocross-linking Panel 2. UV photocross-linked examples of fluorescent barcode strands (left- single coloured cat; right 3 colour orthogonal shapes)
Findings and Conclusion
In conclusion, the authors make an important introduction of a novel high-throughput method of DNA barcoding using patterned photo-crosslinking that requires minimal equipment and provides a versatile experimental design. Furthermore, this approach allows tissue samples to remain intact and for subseuqnrt imaging and experimental steps to occur uninhibited.
LightSeq provides a streamlined and straightforward workflow for scientists to combine in situ imaging and protein staining with sequencing of the same cellular populations, leaving the sample intact for further analysis. We look forward to seeing more exciting work applying the Mightex Polygon to further develop this technique!
Catherine Thomas, PhD Senior Liaison and Development Scientist at Mightex
To read the full publication, please click here.