Abstract
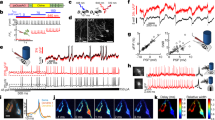
A longstanding goal in neuroscience has been to image membrane voltage across a population of individual neurons in an awake, behaving mammal. Here we describe a genetically encoded fluorescent voltage indicator, SomArchon, which exhibits millisecond response times and is compatible with optogenetic control, and which increases the sensitivity, signal-to-noise ratio, and number of neurons observable several-fold over previously published fully genetically encoded reagents1,2,3,4,5,6,7,8. Under conventional one-photon microscopy, SomArchon enables the routine population analysis of around 13 neurons at once, in multiple brain regions (cortex, hippocampus, and striatum) of head-fixed, awake, behaving mice. Using SomArchon, we detected both positive and negative responses of striatal neurons during movement, as previously reported by electrophysiology but not easily detected using modern calcium imaging techniques9,10,11, highlighting the power of voltage imaging to reveal bidirectional modulation. We also examined how spikes relate to the subthreshold theta oscillations of individual hippocampal neurons, with SomArchon showing that the spikes of individual neurons are more phase-locked to their own subthreshold theta oscillations than to local field potential theta oscillations. Thus, SomArchon reports both spikes and subthreshold voltage dynamics in awake, behaving mice.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
Computer code used to generate results for this study is available at https://github.com/HanLabBU/somarchon-imaging.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request; raw data essential to the work are available online as Source Data files. Sequences of the reported proteins are available at GenBank at the following accession codes: SomArchon MN091368; SomArchon-P2A-CoChR-KV2.1motif, MN091369.
References
Hochbaum, D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014).
St-Pierre, F. et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 17, 884–889 (2014).
Gong, Y., Wagner, M. J., Zhong Li, J. & Schnitzer, M. J. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 5, 3674 (2014).
Adam, Y. et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019).
Chavarha, M. et al. Fast two-photon volumetric imaging of an improved voltage indicator reveals electrical activity in deeply located neurons in the awake brain. Preprint at https://www.biorxiv.org/content/10.1101/445064v2 (2018).
Abdelfattah, A. S. et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019).
Gong, Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015).
Lou, S. et al. Genetically targeted all-optical electrophysiology with a transgenic Cre-dependent optopatch mouse. J. Neurosci. 36, 11059–11073 (2016).
Jin, X., Tecuapetla, F. & Costa, R. M. Basal ganglia subcircuits distinctively encode the parsing and concatenation of action sequences. Nat. Neurosci. 17, 423–430 (2014).
Shi, L. H., Luo, F., Woodward, D. J. & Chang, J. Y. Neural responses in multiple basal ganglia regions during spontaneous and treadmill locomotion tasks in rats. Exp. Brain Res. 157, 303–314 (2004).
Gritton, H. J. et al. Unique contributions of parvalbumin and cholinergic interneurons in organizing striatal networks during movement. Nat. Neurosci. 22, 586–597 (2019).
Flytzanis, N. C. et al. Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditis elegans neurons. Nat. Commun. 5, 4894 (2014).
Piatkevich, K. D. et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018).
Jin, L. et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75, 779–785 (2012).
Chamberland, S. et al. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6, e25690 (2017).
Zou, P. et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 5, 4625 (2014).
Gong, Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015).
Shemesh, O. A. et al. Temporally precise single-cell-resolution optogenetics. Nat. Neurosci. 20, 1796–1806 (2017).
Baker, C. A., Elyada, Y. M., Parra, A. & Bolton, M. M. L. Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. eLife 5, e14193 (2016).
Daigle, T. L. et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018).
Wu, C., Ivanova, E., Zhang, Y. & Pan, Z. H. rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. PLoS ONE 8, e66332 (2013).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Kravitz, A. V. & Kreitzer, A. C. Striatal mechanisms underlying movement, reinforcement, and punishment. Physiology (Bethesda) 27, 167–177 (2012).
Koós, T. & Tepper, J. M. Inhibitory control of neostriatal projection neurons by GABAergic interneurons. Nat. Neurosci. 2, 467–472 (1999).
Zhou, F. M., Wilson, C. J. & Dani, J. A. Cholinergic interneuron characteristics and nicotinic properties in the striatum. J. Neurobiol. 53, 590–605 (2002).
Bittner, K. C. et al. Conjunctive input processing drives feature selectivity in hippocampal CA1 neurons. Nat. Neurosci. 18, 1133–1142 (2015).
Harvey, C. D., Collman, F., Dombeck, D. A. & Tank, D. W. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature 461, 941–946 (2009).
Adam, Y. et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019).
Hansen, K. R. et al. Mild blast injury produces acute changes in basal intracellular calcium levels and activity patterns in mouse hippocampal neurons. J. Neurotrauma 35, 1523–1536 (2018).
Dombeck, D. A., Khabbaz, A. N., Collman, F., Adelman, T. L. & Tank, D. W. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron 56, 43–57 (2007).
Fields, I. E. et al. Noninvasive deep brain stimulation via temporally interfering electric fields. Cell 169, 1029–1041.e16 (2017).
Kalmbach, A. S. & Waters, J. Brain surface temperature under a craniotomy. J. Neurophysiol. 108, 3138–3146 (2012).
Podgorski, K. & Ranganathan, G. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016).
Arias-Gil, G., Ohl, F. W., Takagaki, K. & Lippert, M. T. Measurement, modeling, and prediction of temperature rise due to optogenetic brain stimulation. Neurophotonics 3, 045007 (2016).
Stujenske, J. M., Spellman, T. & Gordon, J. A. Modeling the spatiotemporal dynamics of light and heat propagation for in vivo optogenetics. Cell Rep. 12, 525–534 (2015).
Christie, I. N. et al. fMRI response to blue light delivery in the naïve brain: implications for combined optogenetic fMRI studies. Neuroimage 66, 634–641 (2013).
Dell, R. B., Holleran, S. & Ramakrishnan, R. Sample size determination. ILAR J. 43, 207–213 (2002).
Acknowledgements
We thank T. Ta for help with histology; M. Murdock for help with mouse work; N. Pak, A. Badon and J. Mertz for help with assembling the in vivo imaging setup; and Y. Bando for helping with the Supplementary Video. E.S.B. acknowledges funding from Edward and Kay Poitras, NIH Director’s Pioneer Award 1DP1NS087724, NIH 1R01GM104948, NIH 1R01EB024261, NIH 1R01DA045549, NIH 1R01MH114031, Charles Hieken, NIH 1R01NS102727, John Doerr, NSF Grant 1734870, the HHMI-Simons Faculty Scholars Program, Human Frontier Science Program RGP0015/2016, NIH 1R43MH109332, US Army Research Laboratory and the US Army Research Office under contract/grant number W911NF1510548, NIH 2R01DA029639, and NSF CBET 1344219. X.H. acknowledges funding from the NIH Director’s Office (1DP2NS082126), NINDS (1R01NS109794-01, 1R01NS087950-01), NSF CBET-1848029, the Grainger Foundation, the Pew Foundation, and Boston University Biomedical Engineering Department. S.B. and S.N.S. acknowledge funding from the NIH/NIGMS T32 Quantitative Biology and Physiology Fellowship (GM008764) through the Boston University Biomedical Engineering Department. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.D.P. and E.S.B. initiated the project. K.D.P., S.B., H.T., S.N.S., X.H., and E.S.B. designed all in vivo experiments and interpreted the data. K.D.P. developed SomArchon and together with E.E.J., O.A.S., and E.C. characterized all constructs in cultured cells. K.D.P., V.G.L.-H., D.P., C.S., Z.F., and B.L.S. performed characterization of SomArchon in acute brain slices. S.B., S.N.S. and H.J.G. performed all mouse surgeries for in vivo experiments. M.F.R. assisted on imaging setups. K.D.P, S.B., H.T., and S.N.S performed all in vivo imaging experiments and analysed all in vivo imaging data. K.D.P., S.B., H.T., S.N.S, X.H. and E.S.B. wrote the paper with contributions from all of the authors. E.S.B. and X.H. oversaw all aspects of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Kenneth Harris, Lin Tian, Christian Wilms and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Expression of Archon1 and SomArchon in mouse brain.
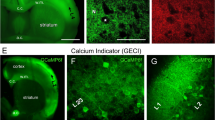
a–d, Representative images of mouse brain slices expressing Archon1 (a, b) and SomArchon (c, d) (CAG promoter, via IUE) imaged with a wide-field microscope with 10× (a, c) and 40× (b, d) objective lenses (from n = 7 slices from 2 mice each). e, f, Normalized EGFP fluorescence along white arrows shown in b, d, respectively. Black dots correspond to resolvable cells. g, Number of resolvable cells per FOV for brain slices expressing Archon1 or SomArchon (2.4 ± 2.5 and 22 ± 9 neurons per FOV (350 × 415 μm2) for Archon1 and SomArchon, respectively). Mean ± s.d.; n = 7 slices from 2 mice each; box plots as in Fig. 1. Further confocal analysis with larger FOVs of 500 × 500 × 50 μm3 revealed that SomArchon can resolve around 15 times more neurons in the cortex than Archon1 (n = 4, 8, 9, 11, 11, 18, and 20 neurons from 7 slices for Archon1, versus n = 180, 187, and 137 neurons from 3 slices for SomArchon). h, Representative confocal images of neurons in cortex layer 2/3 (left), hippocampus (middle), and striatum (right) expressing Archon1 (top) and SomArchon (bottom). i–k, EGFP fluorescence along a neurite, normalized to soma, for neurons expressing Archon1 (left) or SomArchon (right) in cortex layer 2/3 (i, n = 39 and 37 neurites from 10 cells from 2 mice each), hippocampus (j, n = 20 and 34 neurites from 9 and 17 cells from 2 mice each), and striatum (k, n = 17 and 20 neurites from 7 cells from 2 mice each). Box plots as in Fig. 1. *P < 0.002 compared to Archon1 at corresponding position away from the soma; n.s., not significant. Two-sample Kolmogorov–Smirnov test, see Supplementary Table 2. l–u, Representative confocal fluorescence images of brain slices expressing Archon (left) or SomArchon (right) via IUE (l–q) or AAV injection (r–u) in cortex layer 2/3 (n, o; n = 8 slices from 2 mice), hippocampus (p, q; n = 8 slices from 2 mice), and striatum (t, u; n = 6 slices from 2 mice). Green, EGFP; magenta, Nissl staining. Scale bars, 100 μm (a–d), 50 μm (h, n–q, t, u), 250 μm (l, m, r, s).
Extended Data Fig. 2 Voltage imaging using SomArchon in mouse brain slices.
a–d, Representative fluorescence wide-field images of cortex layer 2/3 neurons expressing SomArchon via AAV transduction (a) or IUE (c) with selected ROIs (bottom), and corresponding fluorescence traces (b, d; n = 6 and 13 slices from 2 and 4 mice for AAV transduction and IUE, respectively). Acquisition rate, 632 Hz (b) or 440 Hz (d). e, f, Representative fluorescence wide-field images of striatal neurons expressing SomArchon via AAV transduction (top) with selected ROIs (bottom) (e), and corresponding fluorescence traces (f; n = 8 slices from 2 mice). Acquisition rate, 733 Hz. g, h, Representative fluorescence wide-field images of hippocampal neurons expressing SomArchon via IUE (top) with selected ROIs (bottom) (g), and corresponding fluorescence traces (h; n = 8 slices from 2 mice). Acquisition rate, 333 Hz. i, j, Fluorescence wide-field images of thalamus neurons expressing SomArchon (top) via AAV transduction with selected ROIs (bottom) (i), and corresponding fluorescence traces (j; n = 5 slices from 2 mice). Acquisition rate, 333 Hz. Scale bars, 25 µm.
Extended Data Fig. 3 Expression of SomArchon and voltage imaging do not alter membrane properties or cause phototoxicity.
a, Membrane properties of neurons expressing Archon (hashed boxes) or SomArchon (open boxes) in cortex layer 2/3 brain slices (P = 0.8026, 0.8895, and 0.8236 for resistance, capacitance, and resting potential, respectively; two-sided Wilcoxon rank-sum test comparing Archon1 versus SomArchon; n = 8 and 18 cells from 1 and 2 mice for Archon1 and SomArchon, respectively). b, Similar to a but in hippocampus (P = 0.6294, 0.9720, 0.8880, and 0.0037 for resistance, capacitance, resting potential, and FWHM, respectively; two-sided Wilcoxon rank-sum test comparing negative versus SomArchon; n = 8 and 7 cells from 2 mice each for negative and SomArchon for resistance and resting potential; n = 7 and 7 cells from 2 mice each for negative and SomArchon for capacitance; n = 7 and 8 cells from from 2 mice each for negative and SomArchon for FWHM). c, Similar to a but in striatum (P = 0.7380, 0.8357, 0.7751, and 0.0931 for resistance, capacitance, resting potential and FWHM, respectively; two-sided Wilcoxon rank-sum test comparing negative and SomArchon; n = 7 and 6 cells from 2 mice each for negative and SomArchon for resistance and capacitance; n = 6 and 7 cells from 2 mice each for negative and SomArchon for resting potential; n = 6 and 6 cells from 2 mice each for negative and SomArchon for FWHM). d, Changes in relative ROS concentration (normalized to that before illumination) over time in negative (solid line) and SomArchon-expressing (dashed line) cultured mouse neurons under various illumination protocols. e, Maximal increase in ROS concentration during continuous illumination for conditions performed in d (n = 45, 24, and 8 negative neurons from 2, 2, and 1 cultures for 390/22 nm, 475/36 nm, and 637 nm illumination, respectively; n = 24 SomArchon-expressing neurons for 637 nm illumination from 1 culture). f, Cell death for negative (solid line) and SomArchon-expressing (dashed line) cultured neurons at DIV 14–18 under various illumination protocols (n = 45, 35, 91, 40, and 27 neurons from 2, 1, 2, 1, and 1 cultures, respectively, for 390/22 nm at 2.8 mW mm−2, 390/22 nm at 5.5 mW mm−2, 475/36 nm at 12 mW mm−2, 475/36 nm at 25 mW mm−2, and 637 nm at 1,500 mW mm−2 illumination). g, Bright-field and fluorescence images of representative neurons expressing SomArchon before and after 10 min of continuous 637-nm laser illumination at 1,500 mW mm−2, followed by 10 min in darkness (93% of imaged cells did not exhibit noticeable changes in morphology; n = 27 cells from 1 culture; non-illuminated cells did not show any changes in morphology; n = 10 cells from 1 cultures). Scale bar, 50 μm. h, Representative SomArchon fluorescence trace from neuron co-expressing SomArchon and CoChR-Kv2.1motif. i, Normalized spike rates (to initial value) elicited by blue light illumination dropped after 300 s of continuous recording, owing to decrease in spike amplitude as a result of photobleaching (n = 10 neurons from 1 culture; plotted as mean ± s.d.). j, Normalized (to initial value) FWHM of spikes elicited by continuous light exposure as in h. Box plots as in Fig. 1.
Extended Data Fig. 4 SomArchon expression in vivo does not cause gliosis.
SomArchon was expressed in the mouse brain by AAV2.9-Syn-SomArchon-P2A-CoChR-KV2.1motif injection into the cortex in P0 Swiss Webster mice. Brain tissues were analysed 63 days after viral injection. Merged fluorescence images from 50-μm-thick coronal sections (i) were visualized via EGFP fluorescence of SomArchon (ii), anti-IBA1 immunofluorescence (iii), and anti-GFAP immunofluorescence (iv; n = 4 slices from 2 mice). a, Expression throughout the coronal section. b, Zoomed-in view of the virally injected area (high-expression cortex). c, Zoomed-in view of the non-injected contralateral hemisphere (low-expression cortex). The commonly used glial and microglial markers GFAP and IBA1 appeared similarly in both hemispheres, suggesting that expression of SomArchon did not cause gliosis. Scale bars, 1 mm (a), 250 µm (b, c).
Extended Data Fig. 5 Side-by-side comparison of next-generation voltage indicators in mouse brain slices.
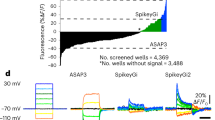
a, Representative fluorescence images of mouse cortex layer 2/3 neurons expressing ASAP3-KV2.1motif (ASAP3-Kv), Ace2N-HaloTag-KGC-ER2-KV2.1motif (Voltron-ST/JF525), QuasAr3-PP-mCitrine-KV2.1motif-ER2 (QuasAr3-s), and paQuasAr3-PP-mCitrine-KV2.1motif-ER2 (paQuasAr3-s). ASAP3-Kv, QuasAr3-s and paQuasAr3-s were visualized via cpGFP, mCitrine, and mCitrine fluorescence, respectively, using laser excitation at 488 nm and emission at 525/50 nm under a confocal microscope. Voltron-ST/JF525 was visualized via JF525 fluorescence using LED excitation at 510/25 nm and emission at 545/40 nm under a wide-field microscope. Scale bar, 50 µm. b, Single-trial optical recordings of ASAP3-Kv (green) and Voltron-ST/JF525 (orange) fluorescence responses during neuronal activity evoked with 4-aminopyridine, and QuasAr3-s (blue), paQuasAr3-s (brown), and SomArchon (red) fluorescence responses during neuronal activity evoked with CoChR-mTagBFP2-KV2.2motif. Acquisition rate, about 500 Hz. Blue light pulses (470/20 nm, 2 ms per pulse, 10 Hz; vertical blue bars) were used to activate CoChR to evoke spiking. c, d, Quantification of ΔF/F (c) and SNR (d) per action potential across all recordings (n = 18, 14, 9, 13, and 14 neurons from 1, 2, 2, 2, and 2 mice for ASAP3-Kv, Voltron-ST/JF525, QuasAr3-s, paQuasAr3-s, and SomArchon, respectively). Box plots as in Fig. 1. *P < 0.01, Wilcoxon rank-sum test; see Supplementary Table 2 for statistics. e, Photobleaching curves of ASAP3-Kv, Voltron-ST/JF525, and SomArchon under continuous illumination (n = 11, 8, and 17 neurons from 1 culture, respectively).
Extended Data Fig. 6 SomArchon enables both local dendritic and population imaging of neurons in multiple brain regions in vivo.
a, Fluorescence images of selected FOV in motor cortex (left) with selected ROIs corresponding to somas of 3 neurons (right) (n = 1 FOV from 1 mouse). Scale bar, 50 μm. b, Representative fluorescence traces from a with detected spikes (black ticks). c, Fluorescence image of a hippocampal neuron expressing SomArchon with ROIs selected at the soma and on 4 proximal dendrites (n = 1 neuron from 1 mouse). Scale bar, 20 μm. d, Optical voltage traces from the selected ROIs shown in c. e, Fluorescence image of a striatal neuron expressing SomArchon with ROIs selected at the soma and on 3 proximal dendrites (n = 1 neuron from 1 mouse). Scale bar, 20 μm. f, Optical voltage traces from the selected ROIs shown in e. Black arrows in d, f highlight instances in which dendritic voltages differed visibly from those on the soma. g–k, In vivo population voltage imaging in the hippocampus CA1 region (n = 14 FOVs from 3 mice). g, i, Average intensity projection image for each video (top), with identified ROIs (bottom). h, j, Optical voltage traces for each neuron shown in g, i, respectively, with colours matching corresponding ROI colours. Panels show 1.2 s of simultaneously recorded voltage for all neurons (left), and a period with prominent spikes (right). Image acquisition rate for all recordings, 826 Hz.
Extended Data Fig. 7 Properties of striatal neurons and movement thresholds.
a, Average firing rate, size, and interspike interval (ISI) for 14 neurons recorded in 9 FOV in 2 mice. Cells simultaneously recorded in the same FOV are colour-coded (blue, red and green). Cells in rows with a white background were recorded individually. b, Selected trace from cell 9 exhibiting spike bursting (top), and a zoomed-in view of the boxed region (bottom). A.U., arbitrary unit. Identified spikes are indicated by the marks on top of the trace. c, Single frame images for FOVs with multiple neurons, colour-coded as in a. Scale bars, 20 μm. d, Representative optical traces from two Cre-dependent SomArchon-expressing striatal cholinergic interneurons in a ChAT-Cre mouse (left; scale bar, 20 μm), recorded in 3 sessions, while mouse was awake with head fixed and navigating a spherical treadmill (n = 2 neurons from 1 mouse). Top trace corresponds to top neuron on left; two bottom traces correspond to bottom neuron. Image acquisition rate, 826 Hz. e, Histogram of instantaneous movement speeds for all FOVs shown in Fig. 3 (nine FOVs in two mice). Instantaneous movement speed was calculated as average speed during each 0.5-s time interval. Red line, threshold for low movement speed identification; green line, threshold for high movement speed identification. f, Histogram of instantaneous movement speed for individual FOVs analysed.
Extended Data Fig. 8 In vivo SomArchon performance over time in the striatum and hippocampus of awake mice.
a–h, Average fluorescence intensity, SNR per spike, and firing rates of neurons in the striatum and hippocampus of awake mice, over multiple trials. a–c, In each striatal recording session, we performed 5 trials, each 12-s long, with inter-trial intervals of 30–60 s. Average fluorescence intensity (a) decreased slightly; spike SNR (b) and firing rates (c) remained constant throughout the recording session (repeated-measures analysis of variance (ANOVA), n = 6 neurons in 5 FOVs from 1 mouse). d–h, In each hippocampal recording session, we performed 10 trials, each 6-s long, with inter-trial intervals of 20–30 s. Average fluorescence intensity (d) showed a slight but significant decrease across trials. SNR (e) decreased between the first and second trials but not afterwards, and firing rate (f) remained constant. Spike amplitude (g) fluctuated randomly over trials, and there was a significant increase in baseline noise (h) between the first and second trials (repeated-measures ANOVA; *P < 0.05, post-hoc test: Tukey’s HSD test, n = 16 neurons in 7 FOVs from 4 mice, Supplementary Table 2). Measurements were normalized to the first trial for each neuron. Box plots as in Fig. 4. i–m, A representative continuous optical trace of a hippocampal neuron over 80 s in an awake, head-fixed mouse (i), with zoomed-in views (j–m) at the beginning and end of the recording highlighting comparable firing rates and SNRs (n = 16 neurons in 7 FOVs from 4 mice).
Extended Data Fig. 9 Analysis of LFP and subthreshold membrane voltage oscillation in the hippocampus.
a, Example hippocampal LFP recordings from a session with ten trials, aligned to the onset of an air puff (green shading) directed to one eye in awake, head-fixed mice. b, LFP power spectrum shows strong theta oscillations. Mean ± s.d., n = 10 trials in 1 session. c, Oscillation power at high frequencies (100–250 Hz, red) and at theta frequencies (blue), aligned to puff onset. Each thin line represents an individual recording session, and the thick lines denote means (n = 7 sessions in 4 mice). d, Eye puff evoked a significant increase in LFP power at high frequency, but not at theta frequency (theta frequency P = 0.5966; high frequency P = 0.0004; two-tailed paired Student’s t-test, n = 7 sessions in 4 mice). Box plots as in Fig. 4. e, Fluorescence image of a representative FOV (top) with selected ROIs (bottom). f, Membrane voltage recorded optically (Vmo) from neurons identified in e, and simultaneously recorded LFPs. Black vertical ticks above Vmos denote spikes. Spike–spike coherence values between neurons are shown on the left and Vmo–Vmo theta coherence values are shown on the right. g, Theta-frequency-filtered LFPs and Vmos for the four traces shown in f. Vmo–LFP coherence values are shown on the right. h, Scatter plot of Vmo–Vmo theta frequency coherence and spike–spike coherence from all neuron pairs, fitted with a linear regression (n = 25 pairs, P = 0.08, t-statistic, r2 = 0.12).
Extended Data Fig. 10 Pairwise coherence and correlation measures over spatial distance.
To investigate the potential of background fluorescence signals under wide-field imaging to produce shared crosstalk signals on neuron pairs, we examined the relationship of various coherence and correlation measures between neurons and background fluorescence over spatial distance. a, b, Pairwise coherence at theta frequencies between neurons. Vmo–Vmo coherence did not decrease significantly with spatial distance. (a, n = 25 pairs analysed with spatial distance of 11–66 μm, centre to centre; b, n = 23 pairs within 50 μm of each other; F = 1.44, P = 0.26, one-way ANOVA). c, d, Pairwise Vmo–Vmo coherence at gamma frequencies (30–50 Hz) was not dependent on spatial distance (c, n = 25 pairs; d, n = 23 pairs within 50 μm of each other; F = 2.10, P = 0.13, one-way ANOVA). e, f, Pairwise correlation between neurons did not decrease significantly with spatial distance (e, n = 25 pairs; f, n = 23 pairs within 50 μm of each other; F = 1.00, P = 0.42, one-way ANOVA). g–l, Same analysis as in a–f performed in background doughnut ROIs surrounding each neuron (Methods). Similar to results from neuron pairs, we found that theta frequency coherence between background doughnut ROIs was not dependent on spatial distance (g, n = 23 pairs; h, n = 21 pairs; F = 0.65, P = 0.59, one-way ANOVA), nor was gamma frequency coherence (i, n = 23 pairs; j, n = 21 pairs; F = 1.93, P = 0.16, one-way ANOVA), or the correlation coefficient (k, n = 23 pairs; l, n = 21 pairs; F = 1.02, P = 0.41, one-way ANOVA). m–o, The coherence between neurons and their corresponding doughnuts was not correlated at theta frequency (m), at gamma frequency (n), or for the Pearson correlation coefficients (o). Box plots are as in Fig. 4.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion.
Supplementary Figures
This file contains Supplementary Figures 1 and 2.
Supplementary Tables
This file contains Supplementary Tables 1-4.
Video 1
Fluorescent recording of spontaneous neuronal activity in a hippocampal neuron in an awake mouse, using SomArchon (imaging acquisition at 826 Hz).
Source data
Rights and permissions
About this article
Cite this article
Piatkevich, K.D., Bensussen, S., Tseng, Ha. et al. Population imaging of neural activity in awake behaving mice. Nature 574, 413–417 (2019). https://doi.org/10.1038/s41586-019-1641-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1641-1
This article is cited by
-
High-speed multiplane confocal microscopy for voltage imaging in densely labeled neuronal populations
Nature Neuroscience (2023)
-
Video-based pooled screening yields improved far-red genetically encoded voltage indicators
Nature Methods (2023)
-
High-speed low-light in vivo two-photon voltage imaging of large neuronal populations
Nature Methods (2023)
-
Theta oscillations represent collective dynamics of multineuronal membrane potentials of murine hippocampal pyramidal cells
Communications Biology (2023)
-
Striatal cholinergic interneuron membrane voltage tracks locomotor rhythms in mice
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.